Researchers in Brazil have performed one and silico study to investigate the similarity of viral epitopes between bovine coronavirus (BCoV) and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and they have found several epitopes presenting to B and T lymphocytes divided between two viruses.
The team also found that the cattle density per. 100,000 people showed a negative association with the increase in cases of coronavirus disease 19 (COVID-19), indicating a potential cross-protection mechanism against SARS-CoV-2 from previous BCoV exposure.
 Study: Potential cross-protection against SARS-CoV-2 from previous exposure to bovine coronavirus. Image credit: Studio Romantic / Shutterstock
Study: Potential cross-protection against SARS-CoV-2 from previous exposure to bovine coronavirus. Image credit: Studio Romantic / Shutterstock
A pre-print version of the study has been published on bioRxiv* server while the article is being peer reviewed.
Background
A long history of cattle taming has promoted the scenario of people sharing infectious substances with cattle, for example OC-43 CoV, a human cold virus, believed to have crossed over from cattle to humans in the past.
Coronaviruses are single-stranded RNA viruses belonging to the family Coronaviridae, which are further composed of four genera: Alfacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus. Betacoronavirus includes SARS-CoV-2 and the aforementioned bovine coronavirus (BCoV), the latter being responsible for diarrhea in newborn calves and respiratory infections in calves and cattle.
The team from Brazil assumes that pre-existing immunity from contact with other coronaviruses, such as BCoV, may be able to induce an adaptive immunity that will help reduce the severity and prevalence of SARS-CoV-2 infection. In a quest to find any possible evidence, the team designed this study to search for peptides derived from BCoV proteins that present antigen to T and B cells and show high identity with their SARS-CoV-2 counterparts. .
Examination methods
The peptide sequences of membrane protein (M), nucleocapsid protein (N), spike protein (S) and replicase polyprotein (ORF1ab) belonging to BCoV were evaluated for their T cell reactivity by predicting their binding to human leukocyte antigen class II (HLA II) molecules, as well as B molecules reactivity / binding.
All BCoV peptides above the cut-off thresholds for T cells and B cell binding were analyzed for their identity with the corresponding peptides of human SARS-CoV-2 using multiple sequence alignment. Sequences with more than 80% identity were selected as peptide matches.
The COVID-19 epidemiology was assessed as the slope of the increase in cases per 100,000 people for each city in the Brazilian state of Mato Grosso do Sul (MS) between January 2020 and September 2021. The slope was compared with the number of cattle per. 100,000 people for each municipality in the MS state.
The distance from each municipality to the metropolitan area in the state sub-region and the general efficiency of public expenditure were used as controls and were compared with the slope of COVID-19 cases in a correlation analysis with COVID-19 prevalence.
Survey results
The team found a total of 136, 23, 45, and 709 15-mer peptides that overlapped by 10 amino acids for S, M, N, and ORF1ab proteins, respectively.
Of the peptides that showed values above the thresholds for T or B cell binding, only 2 peptides from protein S, 2 peptides from protein N and 1 peptide from protein M showed at least 80% similarity to SARS-CoV-2 counterpart peptides. None of the peptides from these three proteins (S, N, M) were found to be above the threshold for T and B cells
However, upon analysis of the ORF1ab protein, 107 peptides showed reactivity to either T or B cells above the threshold. From these, the reactivity of 28 peptides was found to be above the threshold for both T cells and B cells.
The team also found that cattle density per. 100,000 people were negatively correlated with the slope of the COVID-19 case increase. The two parameters used as controls for the study showed, as expected, no association with the slope of COVID-19 cases.
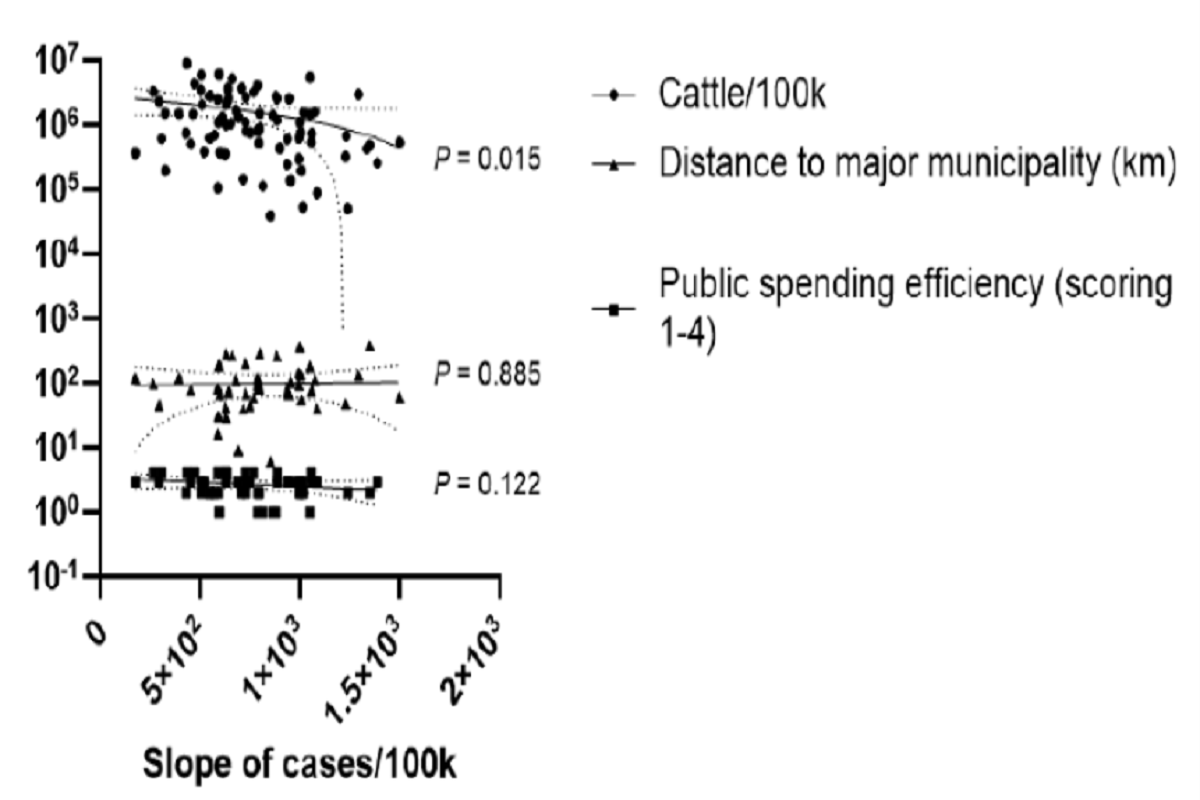 Figure – Linear regression between cattle density and the slope of cumulative COVID-19 random increase in the Brazilian state of Mato Grosso do Sul. Data between Jan / 20 and Sep / 21 were used. Cattle density was calculated as the number of cattle / 100,000 people in the municipality. The distance from the municipality to the large hub town was used to control for lower people the connection to cattle breeding areas. Public spending efficiency was used to check for possible slower responses to the COVID-19 pandemic from cattle breeding municipalities. Analysis by driving test in a linear regression. P-values are displayed for each regression. The dotted lines around the linear regression trend indicate 99% CI.
Figure – Linear regression between cattle density and the slope of cumulative COVID-19 random increase in the Brazilian state of Mato Grosso do Sul. Data between Jan / 20 and Sep / 21 were used. Cattle density was calculated as the number of cattle / 100,000 people in the municipality. The distance from the municipality to the large hub town was used to control for lower people the connection to cattle breeding areas. Public spending efficiency was used to check for possible slower responses to the COVID-19 pandemic from cattle breeding municipalities. Analysis by driving test in a linear regression. P-values are displayed for each regression. The dotted lines around the linear regression trend indicate 99% CI.
Implications
That and silico work with BCoV epitopes reports several epitopes that could be recognized by human T and B lymphocytes and shared with SARS-CoV-2. These epitopes may confer cross-reactivity and are potentially important for the growing immune response to SARS-CoV-2.
The epidemiological data support and silico analysis by showing a possible link between human exposure to cattle and the evolution of the pandemic.
It is possible that the COVID-19 epidemiology was shaped by human exposure to BCoV, just as smallpox was naturally limited by exposure to smallpox, ”the team concludes.
The team finally proposes the use of BCoV as a vaccine candidate against COVID-19.
*Important message
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and therefore should not be considered essential, guide clinical practice / health-related behavior or be treated as established information.
.

Post a Comment